Profile
Kemal Avican is a research fellow working with Maria Fällman in Umeå University Department of Molecular Biology and an IceLab Excellence Center fellow, connected to Stress Response Modeling at IceLab.
Kemal was born and grew up in Nusaybin where one of the most ancient academia, Mor Yakup Church – dated back to 4th century, is located. He then moved to Izmir for his undergraduate education in Biology. During his Molecular Biology master in Gebze Institute of Technology he joined Maria Fällman’s lab at Department of Molecular Biology in Umeå. He then started his PhD studies in the same lab and thereafter a Post-Doc. He joined Mikael Rhen’s lab in Karolinska Institute for his second Post-Doc.
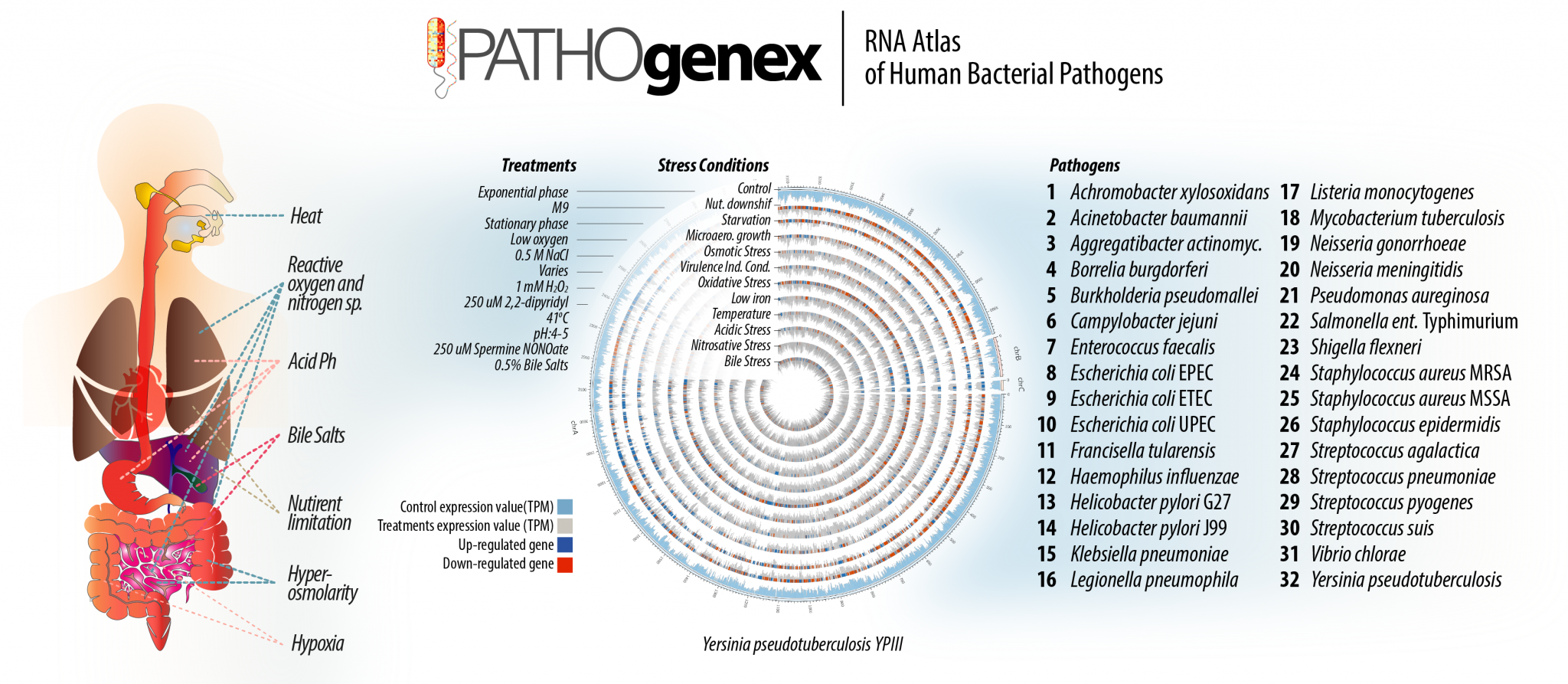
His initial research, focused on Yersinia pseudotuberculosis which is a novel model organism for studying bacterial persistent infections, was expanded with many bacterial pathogens causing worldwide health problems. With collaborative efforts of national and international research groups he could reveal the transcriptomes of 32 human bacterial pathogens under host mimicking stress conditions in PATHOgenex project. He is currently developing novel technics to reach bacterial transcriptomes during infections and deepen PATHOgenex dataset and employ machine learning algorithms in connection with IceLab. His research aims to reveal coordinated regulation and interactions between different gene products involved in a variety of biological processes that are critical for survival of pathogens and identify attractive potential targets for future antimicrobials
Current Projects
Pathogenex
PATHOgenex is a freely available online RNA atlas of 32 human bacterial pathogens exposed to 11 infection relevant stress. The stress exposed bacterial cultures (triplicates) were compared to un-exposed exponentially grown cultures for differential gene expression analysis. The transcriptome data is generated with RNAtag-Seq, which allows multiple library preparation, including rRNA depletion, in one tube.
The Latest Posts
Posts about this Icelabber


