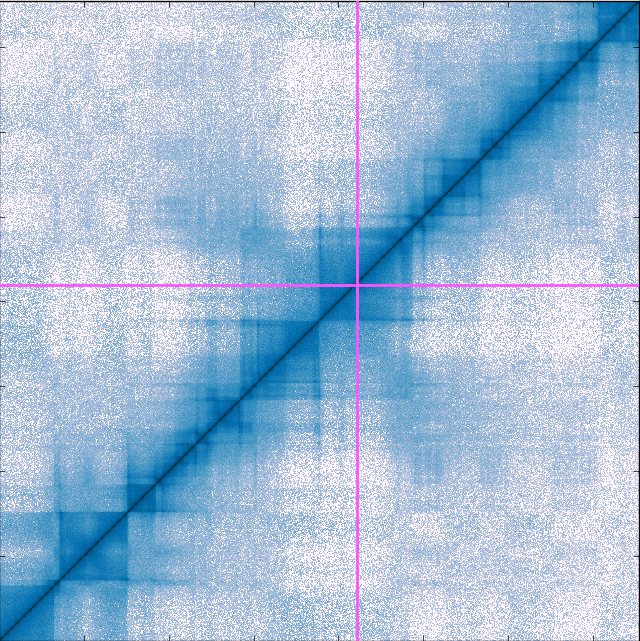
Profile

Associate Professor
Deciphering the life of a cell with physics. Loves crafting new ideas.
Our genetic code has more characters than 850 bible books with about 20,000 genes and millions of regulatory elements. Stored in a two meter long polymer that is folded into a volume smaller than a fog water droplet, how is our DNA organised in 3D to promote important interactions, while avoiding entanglement that erroneously may change how genes turn on and off? Ludvig Lizana runs a research group that applies methods from theoretical physics to figure out how.
Before becoming an associate professor at the physics department in Umeå, and an icelab member, Ludvig worked as a postdoc together with prof. Alexander Grosberg at New York University. Starting out studying electrophoretic motion of large macromolecules (that is, dragging large molecules through a fluid with an electric field), such as DNA, Ludvig got to know about the DNA folding problem which today is a large part of his research. Before moving to New York, Ludvig was a postdoc in prof. Kim Sneppen’s lab, center for models of life, at the Niels Bohr institute (Copenhagen, Denmark). There he worked on various problems such as evolution of metabolic networks in bacteria, protein aggregation in cells, molecular crowding in one dimensional systems (such as protein diffusion along DNA or colloidal particles in nano-channels), and information spreading in social systems. Ludvig did his undergraduate and PhD studies at Chalmers University (Gothenburg, Sweden) where he modeled reaction-diffusion processes in nanofluidic networks.
Current Projects
The Latest Posts
Check out the latest updates by this IceLabber here